Difference Between 16s and 18s rRNA
Brief overview of 16S and 18S rRNA
16s and 18s rRNA molecules can be found in many organisms and play essential roles in protein synthesis as well as being valuable tools in molecular biology research, particularly when studying evolutionary relationships or identifying microbial species.
16S rRNA can be found in bacteria and archaea and is an integral component of their small ribosomal subunit; approximately 1,500 nucleotides long. 16S rRNA’s primary structure exhibits both conserved and variable regions; with conserved regions being essential in upholding structural integrity of a functional ribosome structure and function; therefore its conserved regions have made 16S rRNA an invaluable asset in both phylogenetic studies as well as species identification through 16S gene sequencing techniques.
18S rRNA can be found in various eukaryotes such as plants, animals, fungi and protists and is part of their small ribosomal subunit, consisting of approximately 1,900 nucleotides long. Like 16S rRNA it also features conserved and variable regions to maintain proper structure and function of ribosomes; its gene has also been widely utilized for molecular phylogenetic analyses to understand evolutionary relationships among different eukaryotic organisms.
Although 16s and 18s rRNA both serve similar roles in ribosome assembly and protein synthesis, there are significant variations between them relating to size, sequence, structure and origin; prokaryotic organisms use 16S while eukaryotes prefer 18S versions.
Understanding the differences and similarities between 16s and 18s rRNA are integral for researchers in various fields such as microbiology, evolutionary biology and environmental science, as they enable precise organism identification as well as provide insight into their evolutionary histories.
Definition of rRNA
Ribosomal ribonucleic acid (rRNA) is an integral part of ribosomes – cell structures responsible for protein production – where amino acids are assembled into proteins through translation. Ribosomal rRNA plays an integral part in this process by helping amino acids form proteins during translation.
rRNA molecules are produced via transcription from DNA in the nucleolus of cells within their nuclei and undergoes an intricate maturation process in which modifications and binding with various ribosomal proteins take place. Once produced, functional ribosomes form from this process with one large and one small subunit holding various amounts of rRNA molecules as well as various proteins found on each subunit ribosomal proteins composing their structure.
rRNA serves an integral function in protein synthesis by acting as an intermediary and catalyzing the peptidyl transferase reaction during protein synthesis. Furthermore, its interactions with transfer RNA (tRNA) and messenger RNA (mRNA) ensure accurate reading and translation of genetic information into protein sequences.
rRNA molecules are highly conserved among various organisms, reflecting its essential function within cells. Different kinds of rRNA exist within various species – 16S in prokaryotes and 18S rRNA in eukaryotes for instance – with conserved regions providing opportunities for phylogenetic analysis as well as species identification.
rRNA plays an essential part of ribosomes by contributing to efficient protein synthesis within cells. Studies of its structure, function, and evolution provide valuable insight into organismal structures and evolutionary relationships.
Importance of rRNA in molecular biology
Ribosomal ribonucleic acid (rRNA) plays an essential role in molecular biology due to its vital function in protein synthesis, as well as being used as an invaluable research tool.
To summarize its significance:
- Protein Synthesis: Ribosomes, the organelles responsible for protein synthesis in cells, require ribRNA as they provide structural framework and catalytic activity to transform genetic information from messenger RNA (mRNA) into functional proteins – without which, protein production would become severely limited and compromise cellular functionality.
- Evolutionary Relations: rRNA sequences are highly conserved among organisms and have relatively slow rates of evolution, making their comparison between species possible, which allows reconstruction of evolutionary relationships as well as creation of phylogenetic trees. Analysis of rRNA has greatly enhanced our knowledge about evolution history and relatedness between organisms.
- Taxonomic Identification: RRNA gene sequences provide an invaluable means of classifying microorganisms. 16s and 18s rRNA genes in bacteria and archaea as well as those present in eukaryotes are frequently targeted through techniques like PCR amplification or DNA sequencing for accurate species identification, even for strains that might otherwise prove difficult or impossible to cultivate or identify. Together these techniques collectively known as “rRNA gene sequencing” allow accurate species recognition even with uncultivable or difficult-to-identity microorganisms.
- Environmental Studies: rRNA analysis has revolutionized environmental microbiology. By extracting and sequencing rRNA from environmental samples, researchers can quickly and easily assess microbial diversity and community structure in various ecosystems including soil, water, human gut, etc. This approach, known as metagenomics or environmental DNA sequencing provides invaluable insight into roles and functions played by microorganisms within their natural environments.
- Antibiotics and Drug Development: Many antibiotics target bacterial ribosomes by binding with their rRNA sequence to inhibit protein synthesis, making rRNA study essential in creating antimicrobial agents such as antibiotics or designing therapeutic approaches to combat various diseases. Further investigation can lead to uncovering potential drug targets by studying its structure or function; furthermore rRNA research provides opportunities for understanding antimicrobial strategies against various conditions and designing treatment approaches tailored specifically towards individual disease conditions.
rRNA plays a central role in molecular biology. From its central function in protein synthesis and evolutionary relationships to microbe identification and environmental studies and drug development applications rRNA has proven itself indispensable to expanding our knowledge of biology while improving human health.
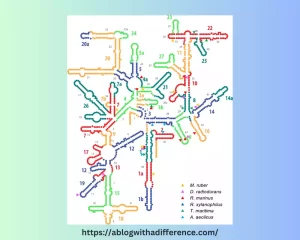
Structure of 16S rRNA
The 16S ribosomal RNA (rRNA) is an integral component of prokaryotic organisms like bacteria and archaea that helps control protein synthesis by providing structural support while decoding genetic information. Structure of 16S rRNA can be divided into two levels of analysis: primary structure and secondary structure.
Primary Structure of 16S rRNA:
- Length: The 16S rRNA molecule can range in length between 1,500 to 2,050 nucleotides depending on species differences, although exact length measurements will often differ significantly between organisms.
- Nucleotide Composition: Nucleic acid sequences contain four nucleotides: Adenine (A), Guanine (G), Cytosine (C) and Uracil (U), which allow species-specific identification and differentiation.
- Conserved and Variable Regions of 16S rRNA: It’s essential for maintaining structural integrity and functionality of ribosomes that there be highly conserved regions among different species that exhibit strong similarity among themselves; such regions help preserve structural integrity of their functions as an organelle. In contrast, variable regions with greater sequence diversity allow us to differentiate closely related organisms more readily.
Secondary Structure :
- Stem-Loop Structures: The 16S rRNA molecule can fold into a complex secondary structure consisting of stem-loop structures created through base pairing between complementary nucleotides within its own molecules forming stem-loops which serve to promote its overall stability and functionality as part of a functional ribosome.
- Helices and Loops: 16S rRNA contains stem-loop structures known as helices that connect via loops; together these helices create three-dimensional structures which serve to bind with proteins or other rRNA molecules to form its three-dimensional architecture and interact with them for binding purposes.
16S rRNA’s secondary structure is highly conserved, especially its core regions essential for assembly and function of ribosomes. This highly conserved secondary structure makes universal primers and probes that target these conserved regions ideal for various uses such as identification of microbes as well as phylogenetic analyses.
16S rRNA provides the necessary framework for ribosome assembly and decoding genetic information during protein synthesis. Furthermore, its conserved and variable regions facilitate species differentiation – making this an indispensable asset in microbe identification and evolutionary studies.
Structure of 18S rRNA
The 18S ribosomal RNA (rRNA) is an integral part of the small ribosomal subunit found in all eukaryotes such as plants, animals, fungi and protists and plays an essential role in protein synthesis by providing structural framework and decoding genetic information. 18S rRNA can be described at two levels of detail; primary structure and secondary structure.
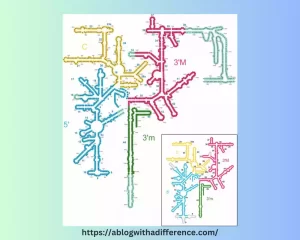
Primary Structure :
- Length of 18S rRNA molecules: (From Wikipedia). Length is typically around 1,900 nucleotides long; this may differ depending on which species it comes from.
- Nucleotide Composition: Each species’ DNA sequence consists of nucleotides like Adenine (A), Guanine (G), Cytosine (C) and Uracil (U), with each specific to that species allowing differentiation and identification.=
- Conserved and Variable Regions of 18S rRNA: It’s well known that 18S rRNA contains conserved regions which are highly similar among different eukaryotic species and essential for maintaining structural integrity and function of ribosomes. Furthermore, variable regions exhibit greater sequence diversity which are useful in distinguishing among closely related organisms.
Secondary Structure :
- Stem-Loop Structures of 18S rRNA: Stem-Loop Structures. The 18S rRNA folds into an intricate secondary structure composed of stem-loop structures formed through base pairing between complementary nucleotides in its molecules; these add stability and functionality to its overall stability and functionality of ribosome.
- Helices and Loops: 18S rRNA’s stem-loop structures form helices connected by loops. Together these helices and loops create three-dimensional structures essential to binding with other ribosomal components such as proteins or other rRNA molecules and for their interaction and binding with one another.
18S rRNA has an extremely conserved structure, particularly within core regions essential to assembly and function of ribosomes. This conservation makes possible universal primers and probes designed to target conserved regions for various applications including phylogenetic analysis and evolutionary analyses in eukaryotes.
18S rRNA provides the framework necessary for proper assembly of ribosomes and decoding of genetic information during protein synthesis in eukaryotic organisms. Furthermore, its conserved and variable regions enable species differentiation – making 18S rRNA an invaluable asset in evolutionary studies and understanding the diversity among eukaryotic organisms.
Differences between 16s and 18s rRNA
16s and 18s rRNA molecules differ significantly in several respects, from size and presence in various organisms to evolution and molecular biology research applications.
Here are key distinctions between 16s and 18s rRNA molecules:
- 16s and 18s rRNA Size and Sequence Variations:
- 16S rRNA: It is approximately 1,500 nucleotides long in prokaryotes.
- 18S rRNA: It is approximately 1,900 nucleotides long in eukaryotes.
- Sequence variations: The nucleotide sequences of 16s and 18s rRNA differ significantly due to their presence in different organisms and evolutionary divergence.
- 16s and 18s rRNA Presence in Different Organisms:
- 16S rRNA: It is found in prokaryotes, including bacteria and archaea.
- 18S rRNA: It is found in eukaryotes, including plants, animals, fungi, and protists.
- 16s and 18s rRNA Evolutionary Relationships:
- 16S rRNA: It is highly conserved among prokaryotes and provides insights into their evolutionary relationships and phylogeny. It has been extensively used for bacterial species identification and phylogenetic analysis.
- 18S rRNA: It is highly conserved among eukaryotes and is used for studying evolutionary relationships and phylogenetic analysis within this domain. It helps in understanding the diversity and relatedness of eukaryotic organisms.
- 16s and 18s rRNA Specific Applications in Molecular Biology Research:
- 16S rRNA: Due to its presence in prokaryotes, 16S rRNA is widely used for microbial identification, especially in bacterial species identification through 16S rRNA gene sequencing. It is also employed in studying microbial communities, environmental microbiology, and microbiome analysis.
- 18S rRNA: Due to its presence in eukaryotes, 18S rRNA is used for phylogenetic analysis and understanding the evolutionary relationships among different eukaryotic organisms. It is also utilized in environmental studies, such as analyzing eukaryotic diversity in various ecosystems.
While there are differences between 16s and 18s rRNA, both play critical roles in protein synthesis, ribosome assembly, and are valuable tools in molecular biology research. They provide insights into the evolutionary history and relatedness of organisms, allowing for the identification and classification of species and the understanding of microbial diversity in different environments.
Comparison chart between 16s and 18s rRNA
Here’s a comparison chart highlighting the key differences and similarities between 16s and 18s rRNA:
| Aspect | 16S rRNA | 18S rRNA |
|---|---|---|
| Length | Approximately 1,500 nucleotides | Approximately 1,900 nucleotides |
| Presence | Found in prokaryotes | Found in eukaryotes |
| Evolutionary Relationships | Provides insights into prokaryotic evolutionary relationships | Provides insights into eukaryotic evolutionary relationships |
| Conserved Regions | Contains conserved regions crucial for ribosome function | Contains conserved regions crucial for ribosome function |
| Variable Regions | Contains variable regions used for species differentiation | Contains variable regions used for species differentiation |
| Molecular Markers | Used for bacterial species identification and phylogenetic analysis | Used for eukaryotic species identification and phylogenetic analysis |
| Applications | Microbial identification, environmental microbiology, microbiome analysis | Phylogenetic analysis, evolutionary studies, environmental studies |
| Importance | Understanding bacterial diversity and relatedness | Understanding eukaryotic diversity and relatedness |
Similarities between 16s and 18s rRNA
16s and 18s rRNA molecules differ by being found in various organisms as well as having different sizes and sequences.
Nonetheless they share certain similarities:
- Role in Ribosome Assembly: 16s and 18s rRNA molecules play an essential part of ribosomal subunit assembly by providing structural framework for protein assembly into functional ribosomes and their subsequent assembly into functioning ribosomes.
- Conservation of Certain Regions: Both 16s and 18s rRNA molecules contain conserved regions essential to maintaining structural integrity and function of ribosomes, often targeted in molecular biology techniques like PCR amplification or DNA sequencing for identification or phylogenetic analysis.
- Use in Phylogenetic Analysis: 16s and 18s rRNA molecules have long been utilized as tools in phylogenetic studies to better comprehend evolutionary relationships and construct trees containing these sequences across species, helping scientists infer histories of evolution as well as relations among them. Researchers can make discoveries related to evolutionary history as they compare sequences among various organisms using this approach.
- Molecular Markers: 16s and 18s rRNA molecules serve as molecular markers to distinguish and classify microorganisms, providing a means to differentiate closely related species as well as an effective means for their identification. They contain specific regions with variable sequence variations to aid with differentiation between closely related organisms as well as identification purposes.
- Importance in Molecular Biology Research: Both 16s and 18s rRNA molecules play a crucial role in various fields of molecular biology research, from studies related to microbial diversity, environmental microbiology, microbiome analysis and evolution biology – providing valuable insight into organism structure, function and evolution.
No matter their differences in size or presence in different organisms, 16s and 18s rRNA remain key players in protein synthesis and powerful tools for understanding biodiversity and relatedness among organisms.
Conclusion
16s and 18s rRNA play an indispensable part in protein synthesis and ribosome assembly, from prokaryotic organisms through to eukaryotes. Though 16S is found more prevalently across species (it occurs predominantly among bacteria), 18S can also be found among eukaryotic ones and shares many similarities – they both possess conserved regions important for proper functioning and serve as molecular markers to detect species identification, evolutionary relationships, environmental microbiology etc.
They’ve long been utilized as part of molecular biology research to study diversity of microbes present within organisms as well as their evolutionary histories! Understanding both similarities and differences is paramount when utilising their scientific applications effectively while simultaneously providing insights into organism structures as a whole as well as their evolutionary histories.